MoClo Kit¶
An implementation of the original MoClo ToolKit for the Python MoClo library.
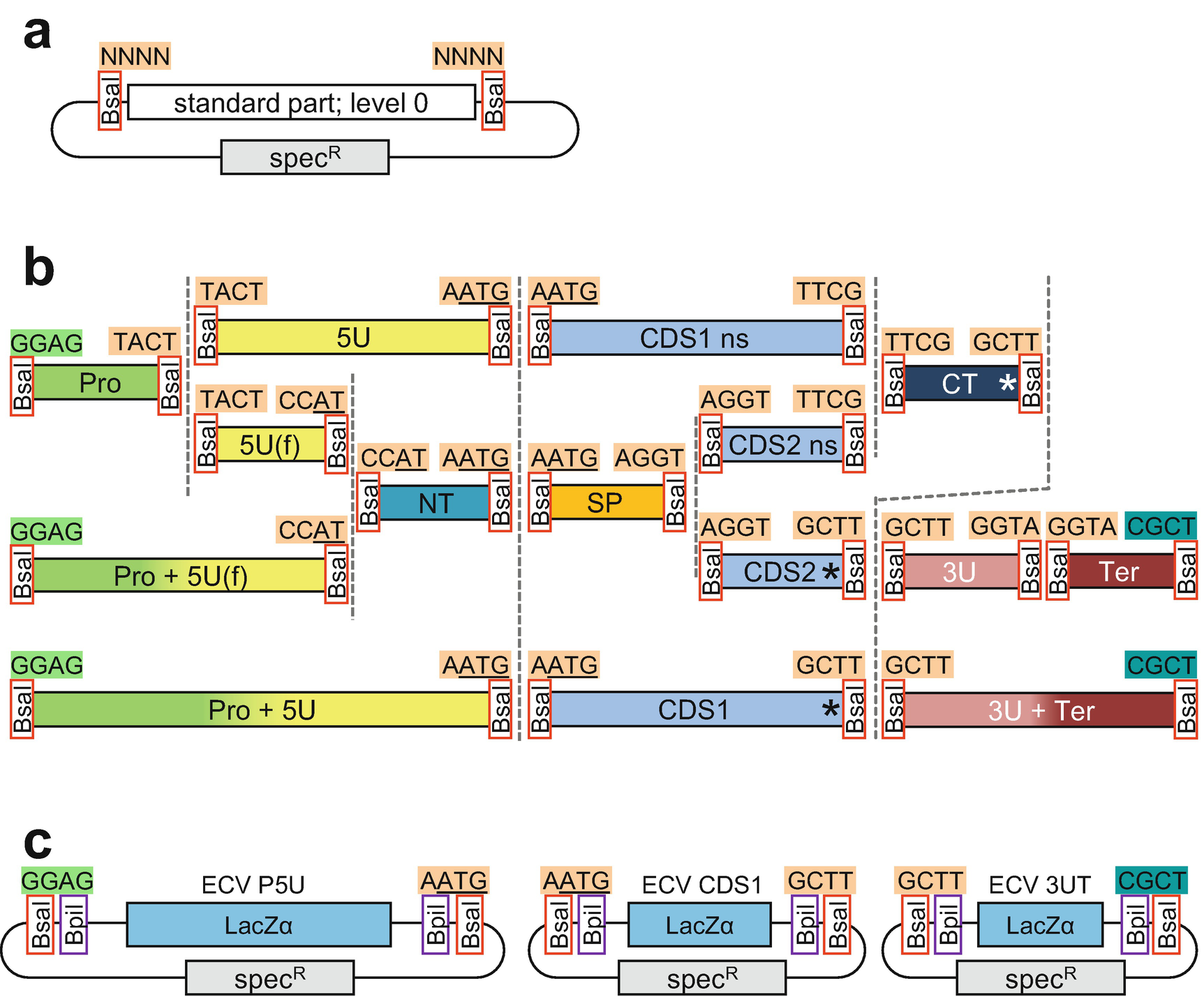
References
Level -1¶
Module¶
- class moclo.kits.moclo.MoCloProduct(Product)[source]¶
An original MoClo product.
- __init__(record)¶
- cutter¶
alias of
Bio.Restriction.Restriction.BpiI
- is_valid()¶
Check if the wrapped record follows the required class structure.
- overhang_end()¶
Get the downstream overhang of the target sequence.
- Returns
the downstream overhang.
- Return type
Seq
- overhang_start()¶
Get the upstream overhang of the target sequence.
- Returns
the downstream overhang.
- Return type
Seq
- classmethod structure()¶
Get the module structure, as a DNA regex pattern.
Warning
If overloading this method, the returned pattern must include 3 capture groups to capture the following features:
The upstream (5’) overhang sequence
The module target sequence
The downstream (3’) overhang sequence
- target_sequence()¶
Get the target sequence of the module.
Modules are often stored in a standardized way, and contain more than the sequence of interest: for instance they can contain an antibiotic marker, that will not be part of the assembly when that module is assembled into a vector; only the target sequence is inserted.
- Returns
the target sequence with annotations.
- Return type
SeqRecord
Note
Depending on the cutting direction of the restriction enzyme used during assembly, the overhang will be left at the beginning or at the end, so the obtained record is exactly the sequence the enzyme created during restriction.
Vector¶
- class moclo.kits.moclo.MoCloEntryVector(EntryVector)[source]¶
A MoClo entry vector.
References
Weber et al., Figure 2A.
- __init__(record)¶
- assemble(module, *modules, **kwargs)¶
Assemble the provided modules into the vector.
- Parameters
module (
AbstractModule) – a module to insert in the vector.modules (
AbstractModule, optional) – additional modules to insert in the vector. The order of the parameters is not important, since modules will be sorted by their start overhang in the function.
- Returns
the assembled sequence with sequence annotations inherited from the vector and the modules.
- Return type
SeqRecord- Raises
DuplicateModules – when two different modules share the same start overhang, leading in possibly non-deterministic constructs.
MissingModule – when a module has an end overhang that is not shared by any other module, leading to a partial construct only
InvalidSequence – when one of the modules does not match the required module structure (missing site, wrong overhang, etc.).
UnusedModules – when some modules were not used during the assembly (mostly caused by duplicate parts).
- cutter¶
alias of
Bio.Restriction.Restriction.BpiI
- is_valid()¶
Check if the wrapped record follows the required class structure.
- overhang_end()¶
Get the downstream overhang of the vector sequence.
- overhang_start()¶
Get the upstream overhang of the vector sequence.
- placeholder_sequence()¶
Get the placeholder sequence in the vector.
The placeholder sequence is replaced by the concatenation of modules during the assembly. It often contains a dropout sequence, such as a GFP expression cassette that can be used to measure the progress of the assembly.
- classmethod structure()[source]¶
Get the vector structure, as a DNA regex pattern.
Warning
If overloading this method, the returned pattern must include 3 capture groups to capture the following features:
The downstream (3’) overhang sequence
The vector placeholder sequence
The upstream (5’) overhang sequence
- target_sequence()¶
Get the target sequence in the vector.
The target sequence if the part of the plasmid that is not discarded during the assembly (everything except the placeholder sequence).
Level 0¶
Module¶
- class moclo.kits.moclo.MoCloEntry(Entry)[source]¶
An original MoClo entry.
- __init__(record)¶
- cutter¶
alias of
Bio.Restriction.Restriction.BsaI
- is_valid()¶
Check if the wrapped record follows the required class structure.
- overhang_end()¶
Get the downstream overhang of the target sequence.
- Returns
the downstream overhang.
- Return type
Seq
- overhang_start()¶
Get the upstream overhang of the target sequence.
- Returns
the downstream overhang.
- Return type
Seq
- classmethod structure()¶
Get the module structure, as a DNA regex pattern.
Warning
If overloading this method, the returned pattern must include 3 capture groups to capture the following features:
The upstream (5’) overhang sequence
The module target sequence
The downstream (3’) overhang sequence
- target_sequence()¶
Get the target sequence of the module.
Modules are often stored in a standardized way, and contain more than the sequence of interest: for instance they can contain an antibiotic marker, that will not be part of the assembly when that module is assembled into a vector; only the target sequence is inserted.
- Returns
the target sequence with annotations.
- Return type
SeqRecord
Note
Depending on the cutting direction of the restriction enzyme used during assembly, the overhang will be left at the beginning or at the end, so the obtained record is exactly the sequence the enzyme created during restriction.
Vector¶
- class moclo.kits.moclo.MoCloCassetteVector(CassetteVector)[source]¶
A MoClo cassette vector.
References
Weber et al., Figure 4A.
- __init__(record)¶
- assemble(module, *modules, **kwargs)¶
Assemble the provided modules into the vector.
- Parameters
module (
AbstractModule) – a module to insert in the vector.modules (
AbstractModule, optional) – additional modules to insert in the vector. The order of the parameters is not important, since modules will be sorted by their start overhang in the function.
- Returns
the assembled sequence with sequence annotations inherited from the vector and the modules.
- Return type
SeqRecord- Raises
DuplicateModules – when two different modules share the same start overhang, leading in possibly non-deterministic constructs.
MissingModule – when a module has an end overhang that is not shared by any other module, leading to a partial construct only
InvalidSequence – when one of the modules does not match the required module structure (missing site, wrong overhang, etc.).
UnusedModules – when some modules were not used during the assembly (mostly caused by duplicate parts).
- cutter¶
alias of
Bio.Restriction.Restriction.BsaI
- is_valid()¶
Check if the wrapped record follows the required class structure.
- overhang_end()¶
Get the downstream overhang of the vector sequence.
- overhang_start()¶
Get the upstream overhang of the vector sequence.
- placeholder_sequence()¶
Get the placeholder sequence in the vector.
The placeholder sequence is replaced by the concatenation of modules during the assembly. It often contains a dropout sequence, such as a GFP expression cassette that can be used to measure the progress of the assembly.
- classmethod structure()[source]¶
Get the vector structure, as a DNA regex pattern.
Warning
If overloading this method, the returned pattern must include 3 capture groups to capture the following features:
The downstream (3’) overhang sequence
The vector placeholder sequence
The upstream (5’) overhang sequence
- target_sequence()¶
Get the target sequence in the vector.
The target sequence if the part of the plasmid that is not discarded during the assembly (everything except the placeholder sequence).
Parts¶
- class moclo.kits.moclo.MoClo5Uf(MoCloPart, MoCloEntry)[source]¶
An original MoClo 5’UTR part for N-terminal tag linkage.
- class moclo.kits.moclo.MoCloNTag(MoCloPart, MoCloEntry)[source]¶
An original MoClo N-terminal tag part.
- class moclo.kits.moclo.MoCloPro5U(MoCloPart, MoCloEntry)[source]¶
An original MoClo promoter fused with a 5’UTR part.
- class moclo.kits.moclo.MoCloPro5Uf(MoCloPart, MoCloEntry)[source]¶
An original MoClo promoter fused with a 5’UTR for N-terminal linkage.
- class moclo.kits.moclo.MoCloCDS1ns(MoCloPart, MoCloEntry)[source]¶
An original MoClo CDS1 without STOP codon for C-terminal tag linkage.
- class moclo.kits.moclo.MoCloSP(MoCloPart, MoCloEntry)[source]¶
An original MoClo signal peptide part.
- class moclo.kits.moclo.MoCloCDS2ns(MoCloPart, MoCloEntry)[source]¶
An original MoClo CDS2 for C-terminal tag linkage.
- class moclo.kits.moclo.MoCloCTag(MoCloPart, MoCloEntry)[source]¶
An original MoClo C-terminal tag part.
Level 1¶
Module¶
Vector¶
Parts¶
Level M¶
Parts¶
- class moclo.kits.moclo.MoCloLevelMVector(MoCloPart, MoCloDeviceVector)[source]¶
- cutter¶
alias of
Bio.Restriction.Restriction.BpiI
- classmethod structure()[source]¶
Get the part structure, as a DNA regex pattern.
The structure of most parts can be obtained automatically from the part signature and the restriction enzyme used in the Golden Gate assembly.
Warning
If overloading this method, the returned pattern must include 3 capture groups to capture the following features:
The upstream (5’) overhang sequence
The vector placeholder sequence
The downstream (3’) overhang sequence
- class moclo.kits.moclo.MoCloLevelMEndLinker(MoCloPart, MoCloCassette)[source]¶
- classmethod structure()[source]¶
Get the part structure, as a DNA regex pattern.
The structure of most parts can be obtained automatically from the part signature and the restriction enzyme used in the Golden Gate assembly.
Warning
If overloading this method, the returned pattern must include 3 capture groups to capture the following features:
The upstream (5’) overhang sequence
The vector placeholder sequence
The downstream (3’) overhang sequence
Level P¶
Parts¶
- class moclo.kits.moclo.MoCloLevelPVector(MoCloPart, MoCloCassetteVector)[source]¶
- cutter¶
alias of
Bio.Restriction.Restriction.BsaI
- classmethod structure()[source]¶
Get the part structure, as a DNA regex pattern.
The structure of most parts can be obtained automatically from the part signature and the restriction enzyme used in the Golden Gate assembly.
Warning
If overloading this method, the returned pattern must include 3 capture groups to capture the following features:
The upstream (5’) overhang sequence
The vector placeholder sequence
The downstream (3’) overhang sequence
- class moclo.kits.moclo.MoCloLevelPEndLinker(MoCloPart, MoCloEntry)[source]¶
- cutter¶
alias of
Bio.Restriction.Restriction.BsaI
- classmethod structure()[source]¶
Get the part structure, as a DNA regex pattern.
The structure of most parts can be obtained automatically from the part signature and the restriction enzyme used in the Golden Gate assembly.
Warning
If overloading this method, the returned pattern must include 3 capture groups to capture the following features:
The upstream (5’) overhang sequence
The vector placeholder sequence
The downstream (3’) overhang sequence