YTK integration vector¶
In this example, we will be using the moclo library as well as the moclo-ytk extension kit to generate the pre-assembled YTK integration vector (pYTK096) from the available YTK parts, as described in the *Lee et al.* paper
Structure¶
The list of parts, as well as the vector structure, can be found in the Supporting Table S1 from the Lee et al. supplementary materials:

Loading parts¶
We’ll be loading each of the desired parts from the moclo-ytk registry. It is generated from the GenBank distributed with the YTK kits. They can be found on the AddGene YTK page.
[2]:
from moclo.registry.ytk import YTKRegistry
registry = YTKRegistry()
vector = registry['pYTK090'].entity # Part 8a
modules = [registry['pYTK008'].entity, # Part 1
registry['pYTK047'].entity, # Part 234r
registry['pYTK073'].entity, # Part 5
registry['pYTK074'].entity, # Part 6
registry['pYTK086'].entity, # Part 7
registry['pYTK092'].entity] # Part 8b
Checking parts¶
We can use dna_features_viewer to visualize your records before proceeding (for readability purposes, we’ll show the records as linear although they are plasmids):
[3]:
import itertools
import dna_features_viewer as dfv
import matplotlib.pyplot as plt
translator = dfv.BiopythonTranslator([lambda f: f.type != 'source'])
plt.figure(1, figsize=(24, 10))
for index, entity in enumerate(itertools.chain(modules, [vector])):
ax = plt.subplot(2, 4, index + 1)
translator.translate_record(entity.record).plot(ax)
plt.title(entity.record.id)
plt.show()
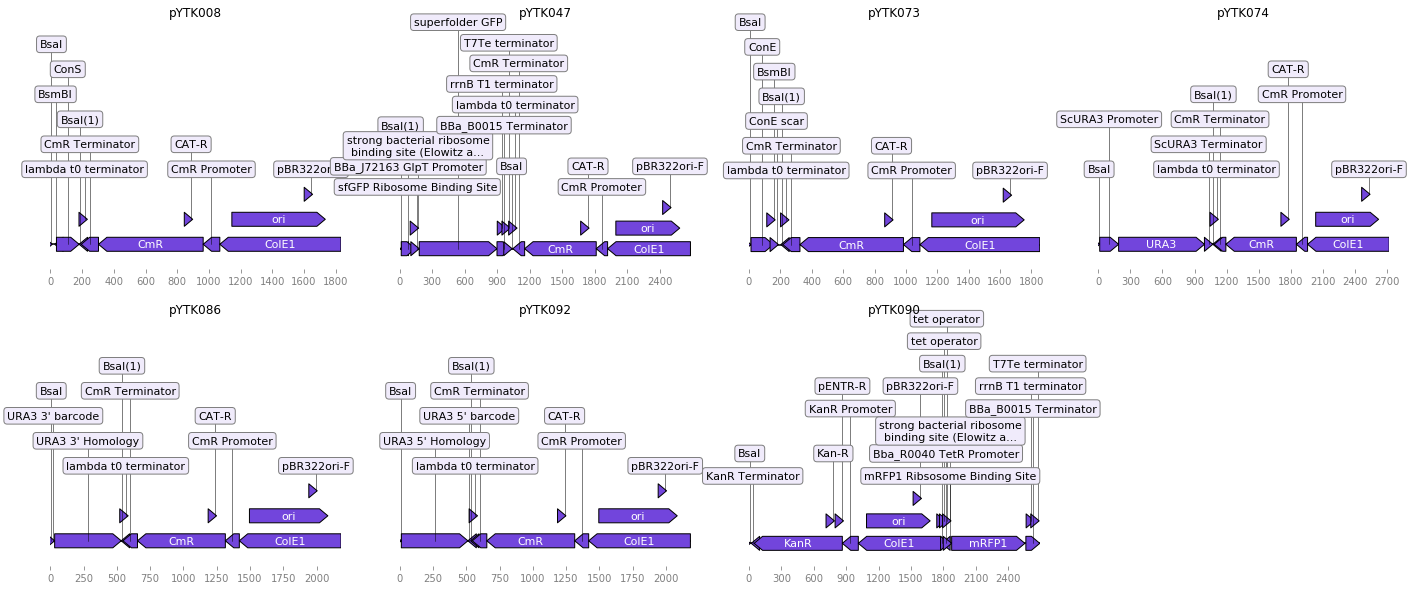
Creating the assembly¶
We use the Part 8a as our base assembly vector, and then assemble all the other parts into that vector:
[4]:
assembly = vector.assemble(*modules)
Rendering the assembly sequence map¶
When creating an assembly, corresponding regions of the obtained sequence will be annotated with the ID of the sequence they come from. We build a simple translator to color the different parts of the plasmid like in the original paper.
With the translator ready, we can display the pre-assembled integration vector assembled by moclo:
[6]:
vec_translator = IntegrationVectorTranslator([lambda f: f.type == 'source'])
vec_translator.translate_record(assembly, dfv.CircularGraphicRecord).plot(figure_width=8)
plt.show()
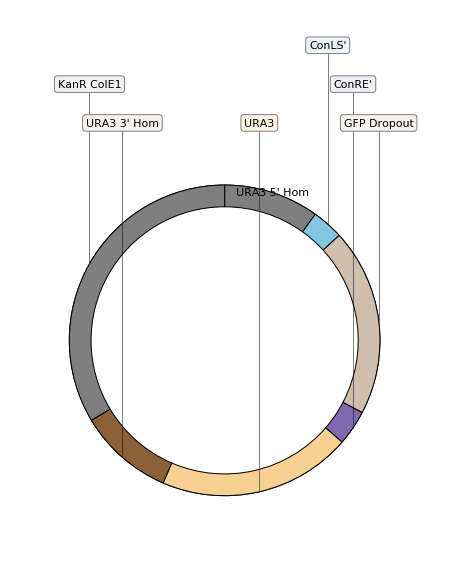
Comparing the assembly to the expected vector¶
Hopefully the obtained assembly should look like the pYTK096 plasmid, distributed with the official YTK parts:
[7]:
plt.figure(3, figsize=(24, 10))
ax = plt.subplot(2, 1, 1)
translator.translate_record(assembly).plot(ax)
plt.title('Assembly')
ax = plt.subplot(2, 1, 2)
translator.translate_record(registry['pYTK096'].entity.record).plot(ax)
plt.title('Expected')
plt.show()
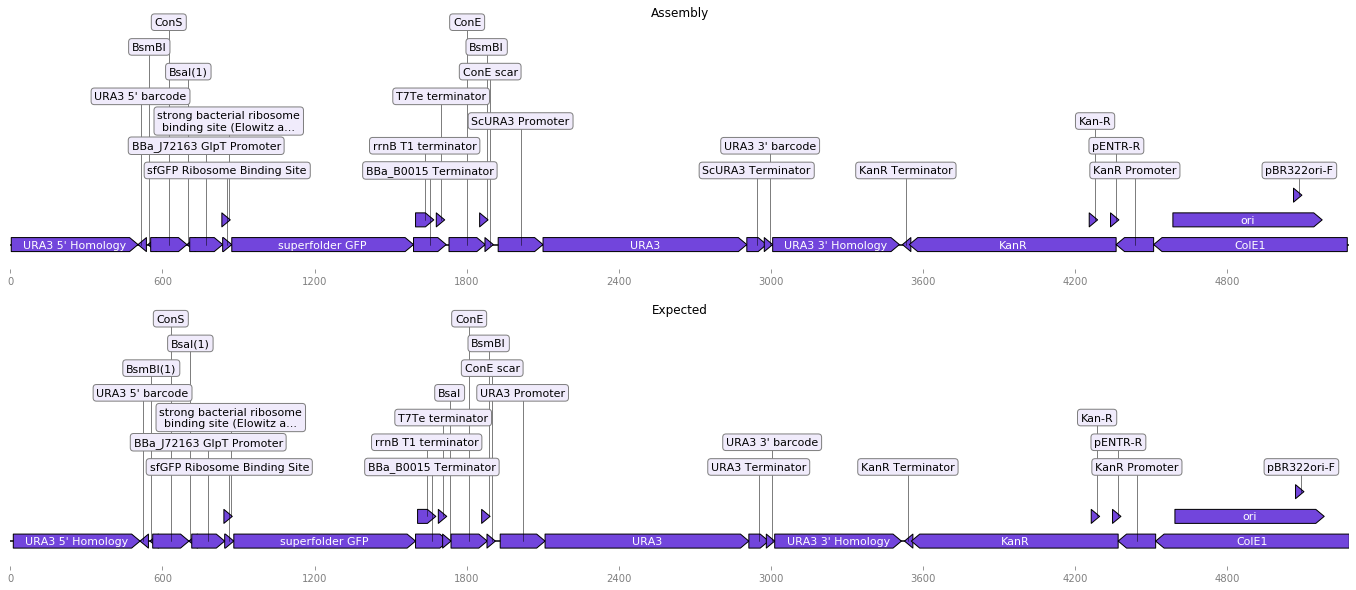
[ ]: